🙋♂️ About Me
I currently have the privilege of working as a Postdoctoral Researcher at the Kather Lab, Else Kroener Fresenius Center for Digital Health, Technical University Dresden, under the supervision of Prof. Jakob Nikolas Kather. Before 2026, I was a PhD student at The School of Computing and Data Science, The University of Hong Kong, advised by Professor Lequan Yu, who direct the MedAI Lab. I also collaborated with Professor Yuanhua Huang closely. I obtained my B.S. degree in Computer Science at Beihang Univerisity in 2021, advised by Professor Guoqi Li and Professor Lei Deng.
My research lies at the intersection of Artificial Intelligence, Healthcare and Biomedical Sciences. I am dedicated to designing advanced machine learning methods and computational algorithms for analysing medical image and biomedical data.
Currently, I mainly focus on deep learning algorithms for Computational Pathology and Spatial Omics Analysis.
🔎 I am happy for collaboration on AI for healthcare research! Please feel free to email me at wqzhao98@conncet.hku.hk.
🔥 News
-
2025.11: 🔥 Thrilled to share that our pathology FM adaptation work, Knowledge-Guided Adaptation of Pathology Foundation Models Effectively Improves Cross-domain Generalization and Demographic Fairness, is accepted in principle by Nature Communications! Congratulats to my colloaborator Yanyan! The paper will be online soon and the codes have been made available!
-
2025.10: 🎉 Started my new position as a Postdoctoral Researcher at Kather Lab. I am super privileged to work under the guidance of Prof. Kather.
-
2025.08: 🔥 Thrilled to share that our paper, Hist2Cell: Deciphering Fine-grained Cellular Architectures from Histology Images, is accepted in principle by Cell Genomics! The codes + datasets have been made available!
-
2025.06: 🎉 Two papers accepted by MICCAI 2025, congratulations to Peixiang and Liting!
-
2025.04: 🔥 Excited to share our latest work FLEX! An amazing work on Pathology Model Adaptation! Check our implementation and let’s train AI to generalize like human doctors among different clinical sites and demographic group!
-
2025.03: 🔥 Our paper TAD-Graph: Enhancing Whole Slide Image Analysis via Task-Aware Subgraph Disentanglement has been accepted by IEEE Transactions on Medical Imaging (TMI)! Check the online publication and the source code is also available now!
-
2025.02: 🔥 Our work From Layers to States: A State Space Model Perspective to Deep Neural Network Layer Dynamics is accepted by ICLR 2025! Check the full paper on Arxiv and see you in Singapore! Official Codes will come soon, stay tuned!
-
2024.12: 🔥 Excited to share our latest work Aligning Knowledge Concepts to Whole Slide Images for Precise Histopathology Image Analysis published on Nature npj Digital Medicine! Check the online publication, and codes + prompts have been made available!
-
2024.12: 🔥 Excited to share our latest ST-Pathology Foundation Model on BRIDGE, a CLIP-based FM pre-trained on over 600,000 image-gene paired profiles from 13 organs with various sequencing techniques! The codes, model weights and collected dataset are online now and the full paper will be released soon, stay tuned!
-
2024.09: 🎉 Our paper Free Lunch in Pathology Foundation Model: Task-specific Model Adaptation with Concept-Guided Feature Enhancement is accepted by NeurIPS 2024! Check the full paper, and codes + prompts have been made available!
-
2024.09: 🔥 Excited to share our latest work Hist2Cell: Deciphering Fine-grained Cellular Architectures from Histology Images. Our model predicts transcriptional cell types from cheap histology images and enable precise survival analysis. Check the full paper on bioRxiv, and codes + datasets have been made available!
-
2024.08: 🎉 Our short paper ST-200K: A Large-scale Paired Dataset for Histology Imaging and Spatial Transcriptomics is accepted by MLCB 2024. A bigger dataset and foundation model are coming soon!
-
2024.06: 🎉 Our paper Unleash the Power of State Space Model for Whole Slide Image with Local Aware Scanning and Importance Resampling is accepted by IEEE Transactions on Medical Imaging (TMI)! Check the full paper and codes.
-
2023.07: 🔥 Our paper ConSlide: Asynchronous Hierarchical Interaction Transformer with Breakup-Reorganize Rehearsal for Continual Whole Slide Image Analysis is accepted by ICCV 2023! Check the full paper, and codes have been made available!
-
2023.06: 🔥 Our paper HIGT: Hierarchical Interaction Graph-Transformer for Whole Slide Image is accepted by MICCAI 2023! Check the full paper, and codes have been made available!
-
2023.01: 🎉 Our paper Transformer-based Multimodal Fusion for Survival Prediction by Integrating Whole Slide Images, Clinical, and Genomic Data is accepted by ISBI 2023! Check the full paper.
-
2022.11: 🔥 Our paper MulGT: Multi-Task Graph-Transformer with Task-Aware Knowledge Injection and Domain Knowledge-Driven Pooling for Whole Slide Image Analysis is accepted by AAAI 2023! Check the full paper, and codes have been made available!
- 2022.08: 🎉 Our short paper Transformer-based Multimodal Fusion for Survival Prediction from Whole Slide Images is accepted by MLCB 2022!
📚 Publications
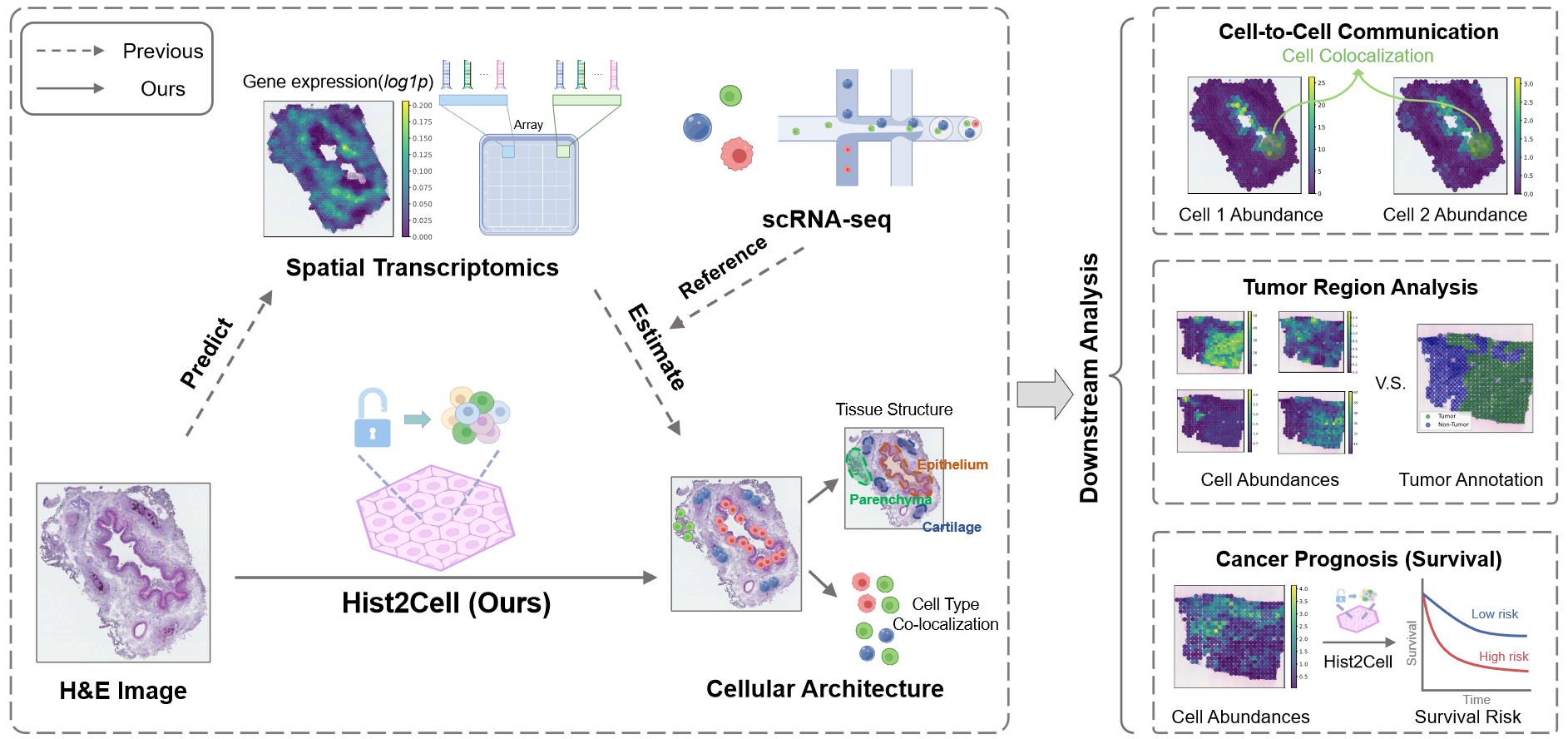
Hist2Cell: Deciphering Fine-grained Cellular Architectures from Histology Images
Weiqin Zhao, Zhuo Liang, Xianjie Huang, Yuanhua Huang, Lequan Yu.
Cell Genomics Accepted in Principle
Developed a one-stage prediction model for identifying spatial transcription-related fine-grained cell types from histology images, enhancing cost-efficient analysis for cancer studies and clinical applications. Achieved accurate cell-type identification and colocalization on external datasets, validating existing biological findings using cost-effective histology images; Enabled large-scale fine-grained cellular analysis on public histology cohorts, producing significant consensus findings that were previously unattainable.
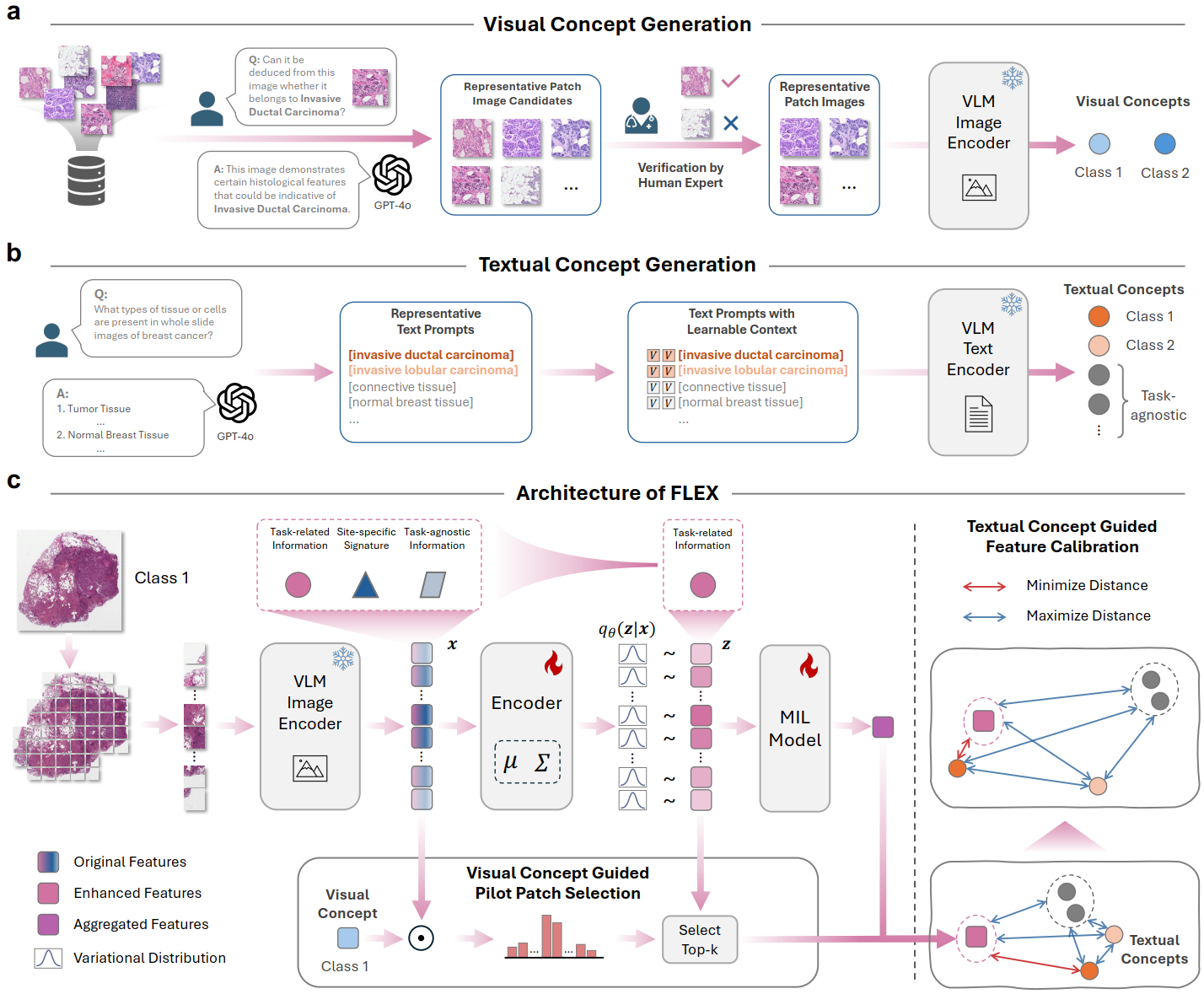
Knowledge-Guided Adaptation of Pathology Foundation Models Effectively Improves Cross-domain Generalization and Demographic Fairness
Yanyan Huang, Weiqin Zhao, Yihang Chen, Yu Fu, Yuming Jiang, Li Liang, Shujun Wang, Lequan Yu.
Nature Communications Accepted in Principle
Proposed FLEX, a novel framework that enhances pathology foundation models to address critical limitations in cross-domain generalization and demographic fairness caused by site-specific signatures and biases. FLEX utilizes a task-specific information bottleneck guided by visual and textual domain knowledge to align features across diverse clinical sites and demographic groups. Evaluated on 16 tasks across 4 TCGA cohorts, FLEX significantly improves out-of-distribution performance, enhances fairness, and shows high versatility with different VLMs and MIL methods, promoting more robust and equitable computational pathology.
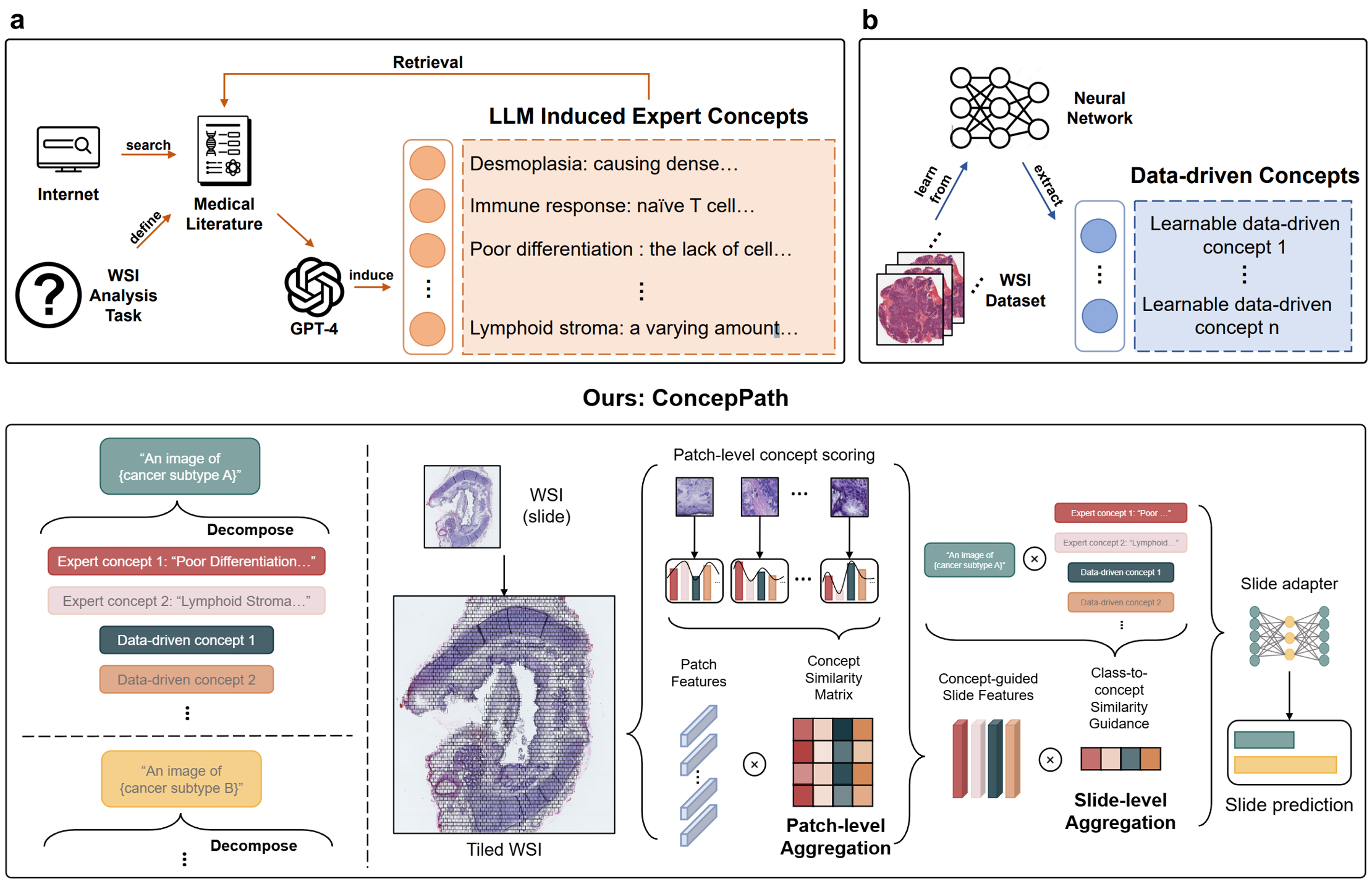
Aligning Knowledge Concepts to Whole Slide Images for Precise Histopathology Image Analysis
Weiqin Zhao, Ziyu Guo, Yinshuang Fan, Yuming Jiang, Maximus Yeung, Lequan Yu.
Nature npj Digital Medicine, 2024
Developed a knowledge concept-based framework for precise Whole Slide Imaging (WSI) analysis by integrating human expert knowledge with data-driven concepts. Employed Large Language Models (LLMs) to derive reliable human expert knowledge from medical literature and align it with histology images using a pathology vision-language model. Achieved enhanced automated diagnostic precision in various cancers.
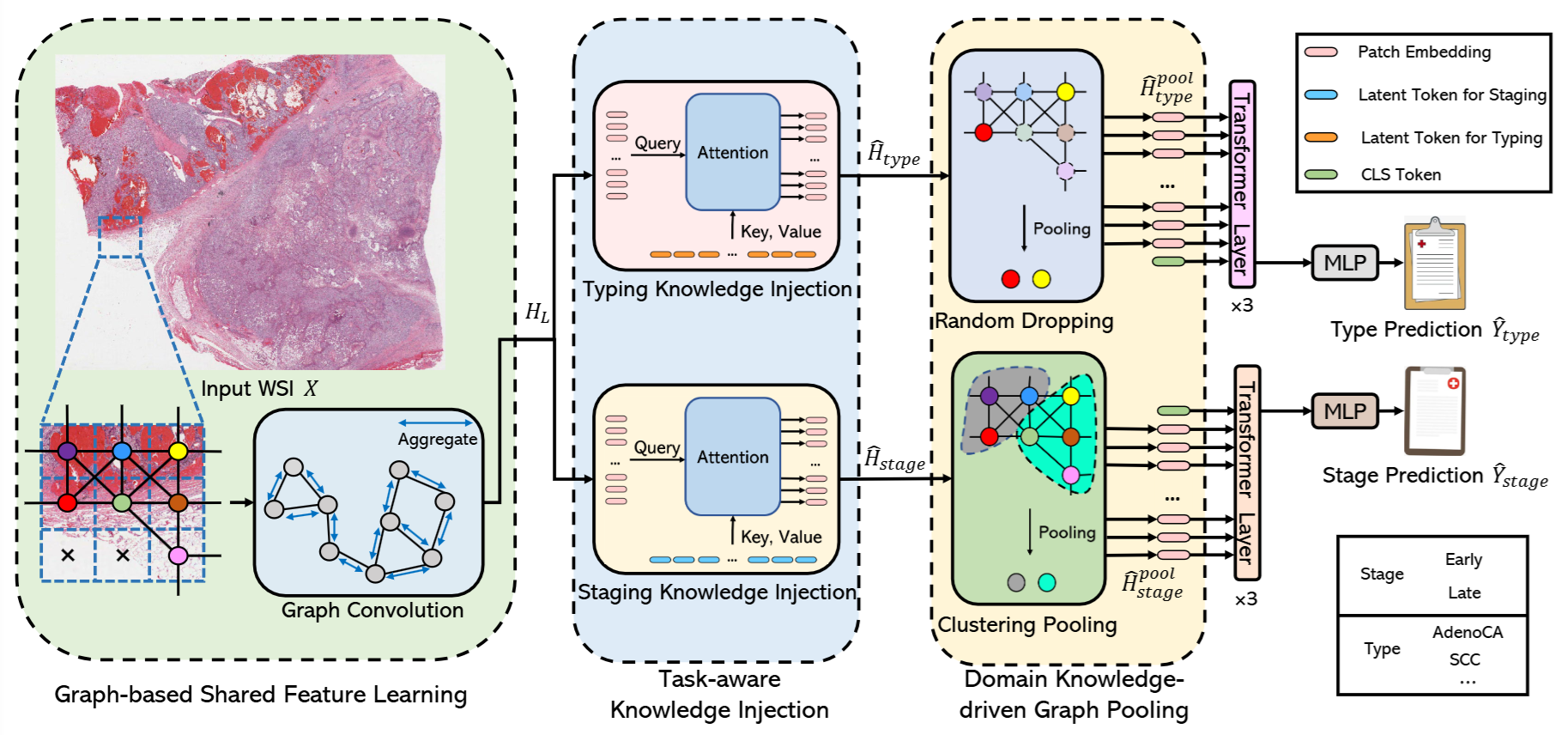
MulGT: Multi-Task Graph-Transformer with Task-Aware Knowledge Injection and Domain Knowledge-Driven Pooling for Whole Slide Image Analysis
Weiqin Zhao, Shujun Wang, Maximus Yeung, Tianye Niu, Lequan Yu.
The AAAI Conference on Artificial Intelligence (AAAI), 2023
This framework leverages Graph Neural Networks and Transformers to learn both task-agnostic local features and task-specific global representations. It incorporates a Task-aware Knowledge Injection module that adapts shared graph embeddings into task-specific feature spaces and a Domain Knowledge-driven Graph Pooling module that uses diagnostic patterns to enhance accuracy and robustness.
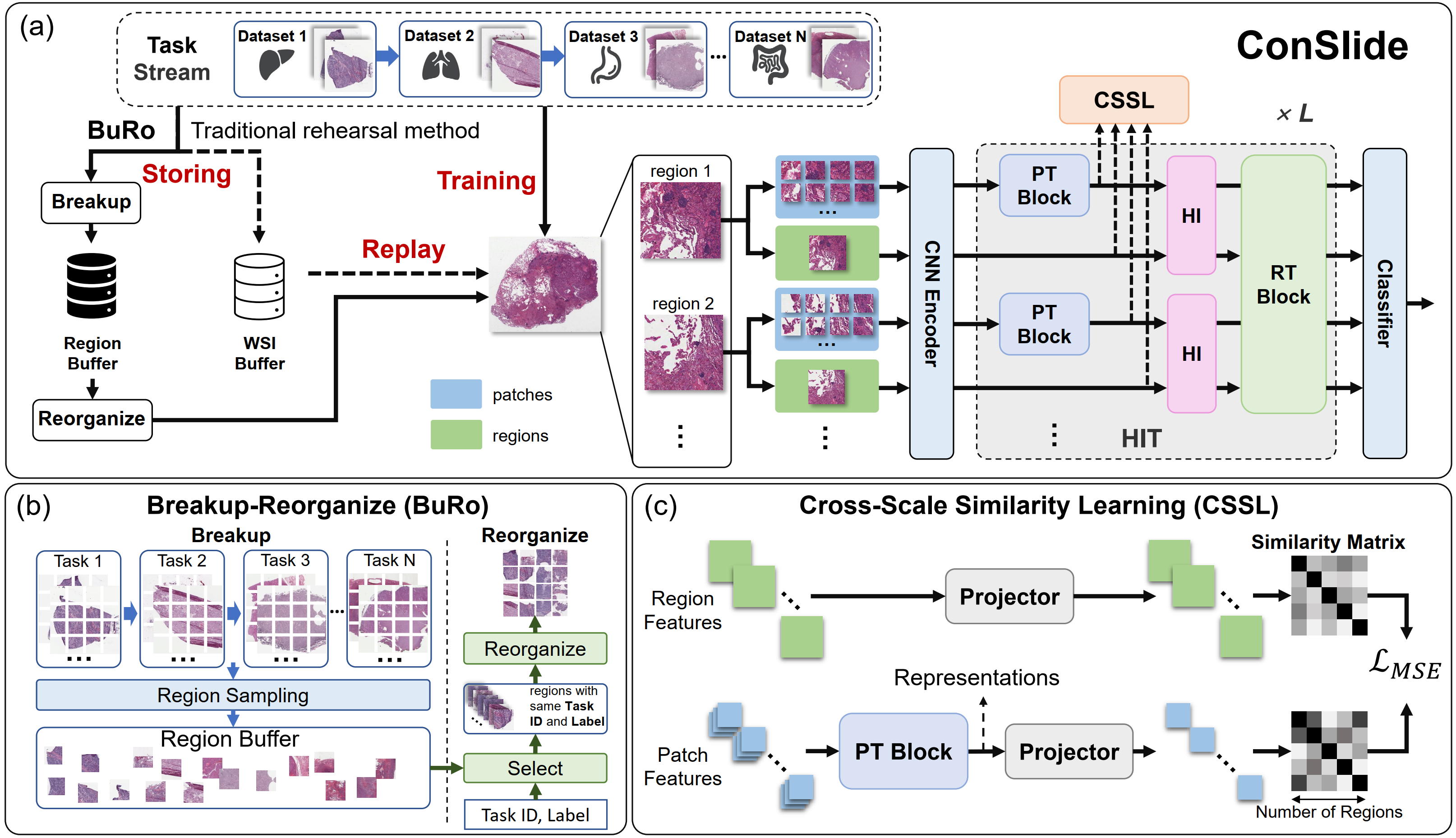
ConSlide: Asynchronous Hierarchical Interaction Transformer with Breakup-Reorganize Rehearsal for Continual Whole Slide Image Analysis
Yanyan Huang$^{*}$, Weiqin Zhao$^{*}$, Shujun Wang, Yu Fu, Yuming Jiang, Lequan Yu.
International Conference on Computer Vision (ICCV), 2023
Propose the first continual learning framework for WSI analysis, named ConSlide, to tackle the challenges of enormous image size, utilization of hierarchical structure, and catastrophic forgetting by progressive model updating on multiple sequential datasets.
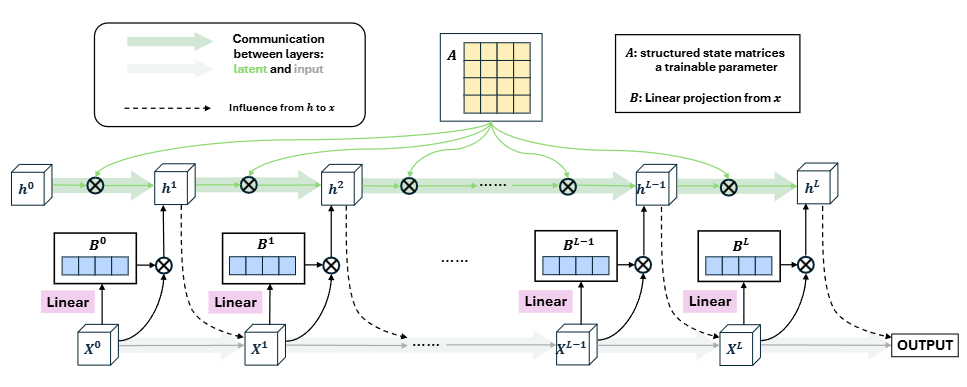
From Layers to States: A State Space Model Perspective to Deep Neural Network Layer Dynamics
Qinshuo Liu$^{*}$, Weiqin Zhao$^{*}$, Wei Huang, Yanwen Fang, Lequan Yu, Guodong Li.
International Conference on Learning Representations (ICLR), 2025
This paper novelly treats the outputs from network layers as states of a continuous process and considers leveraging the state space model (SSM) to design the aggregation of layers in very deep neural networks. Designed a new module called Selective State Space Model Layer Aggregation (S6LA) to combine traditional CNN or ViT within a sequential framework, enhancing its representational capabilities.
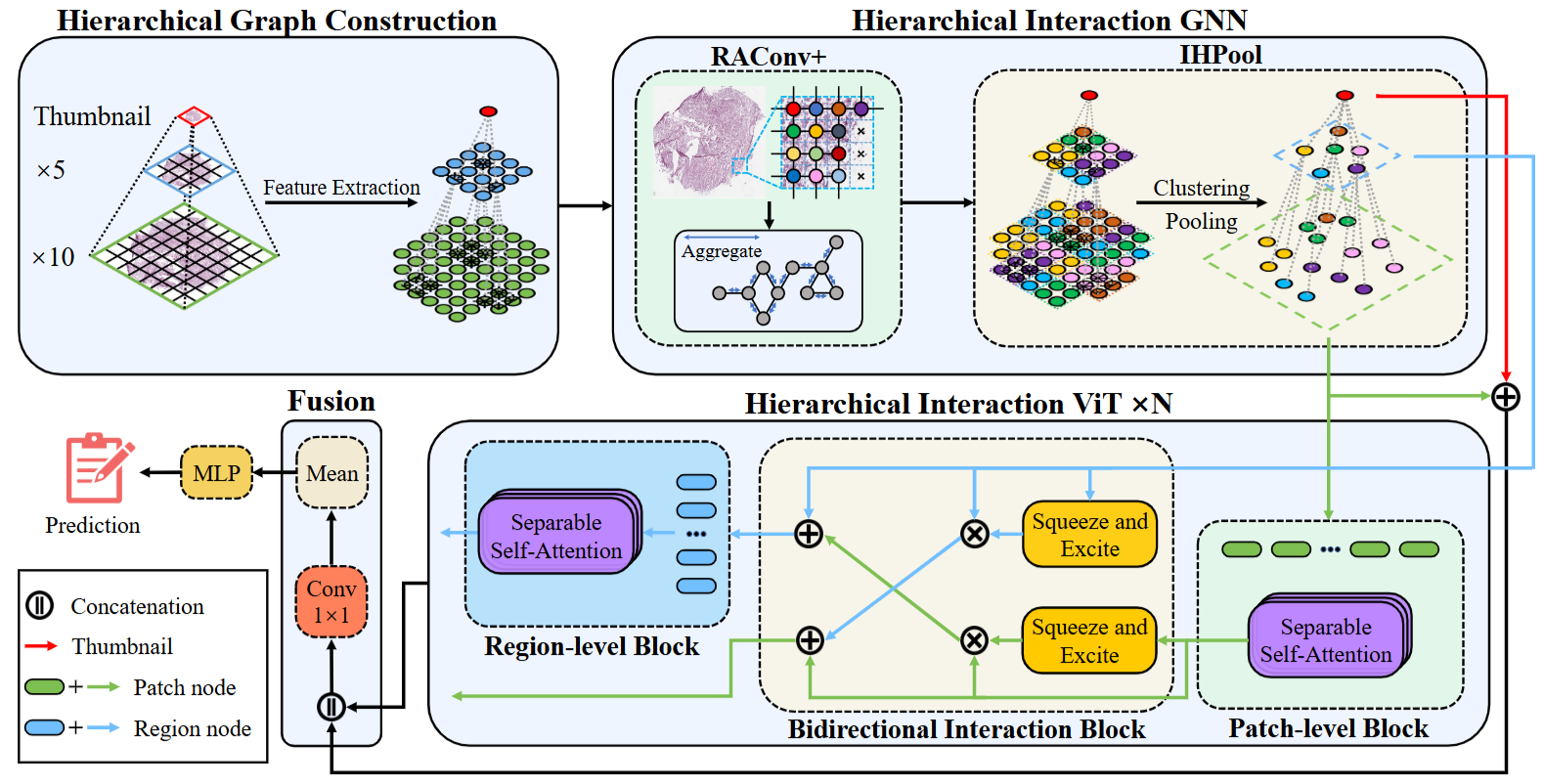
HIGT: Hierarchical Interaction Graph-Transformer for Whole Slide Image
Ziyu Guo$^{*}$, Weiqin Zhao$^{*}$, Shujun Wang, Lequan Yu.
International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), 2023
This paper introduces a novel Hierarchical Interaction GraphTransformer (HIGT) for Whole Slide Imaging (WSI) analysis. Built on Graph Neural Networks and Transformers, HIGT effectively captures both short-range local information and long-range global representations from WSI pyramids. Recognizing the complementary nature of information across different resolutions, we design a Bidirectional Interaction block to enable communication between different levels of the WSI pyramids, enhancing the learning process.
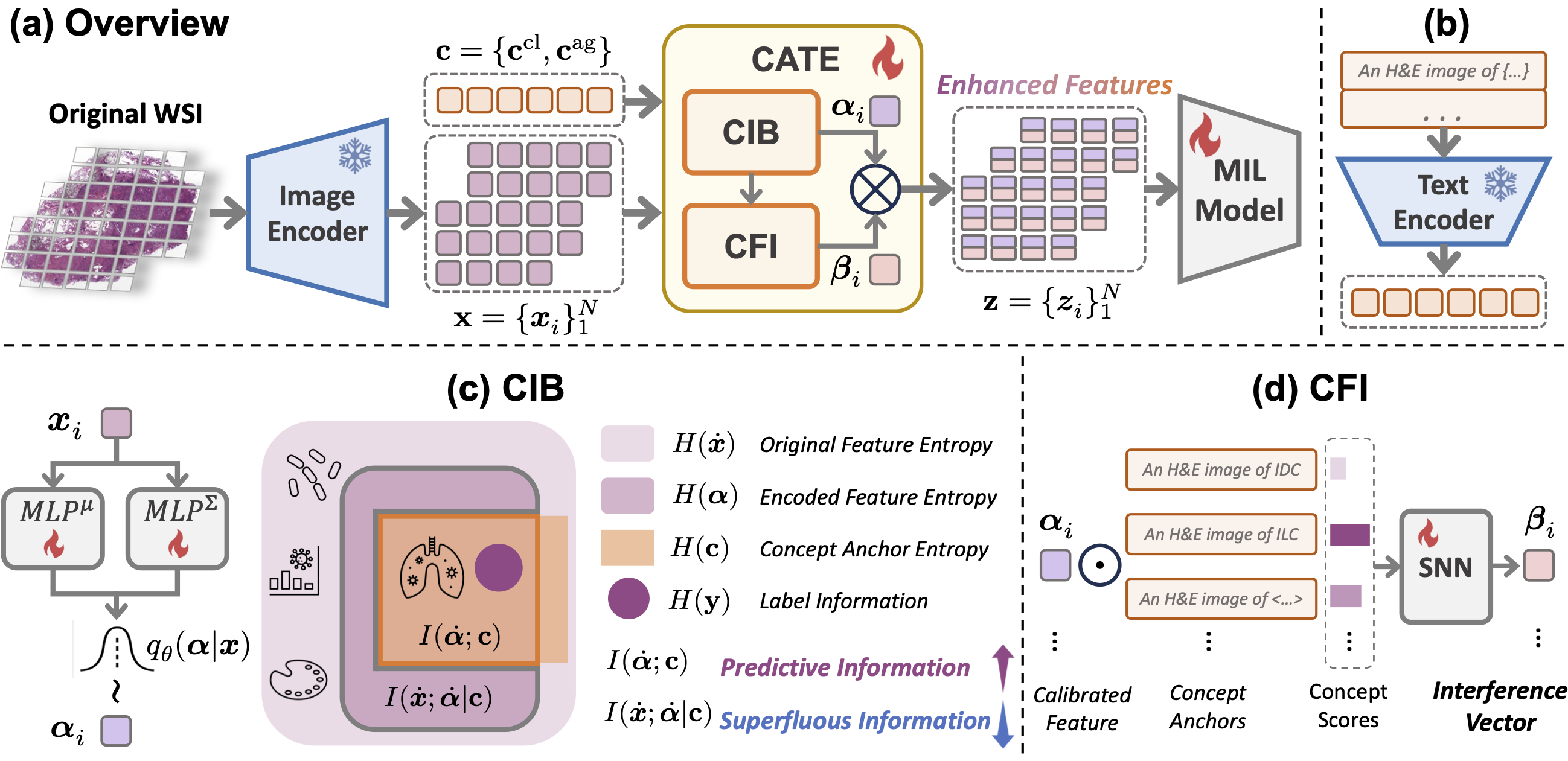
Free Lunch in Pathology Foundation Model: Task-specific Model Adaptation with Concept-Guided Feature Enhancement
Yanyan Huang, Weiqin Zhao, Yihang Chen, Yu Fu, Lequan Yu.
Conference on Neural Information Processing Systems (NeurIPS), 2024
Present Concept Anchor-guided Task-specific Feature Enhancement (CATE), an adaptable paradigm that can boost the expressivity and discriminativeness of pathology foundation models for specific downstream tasks.
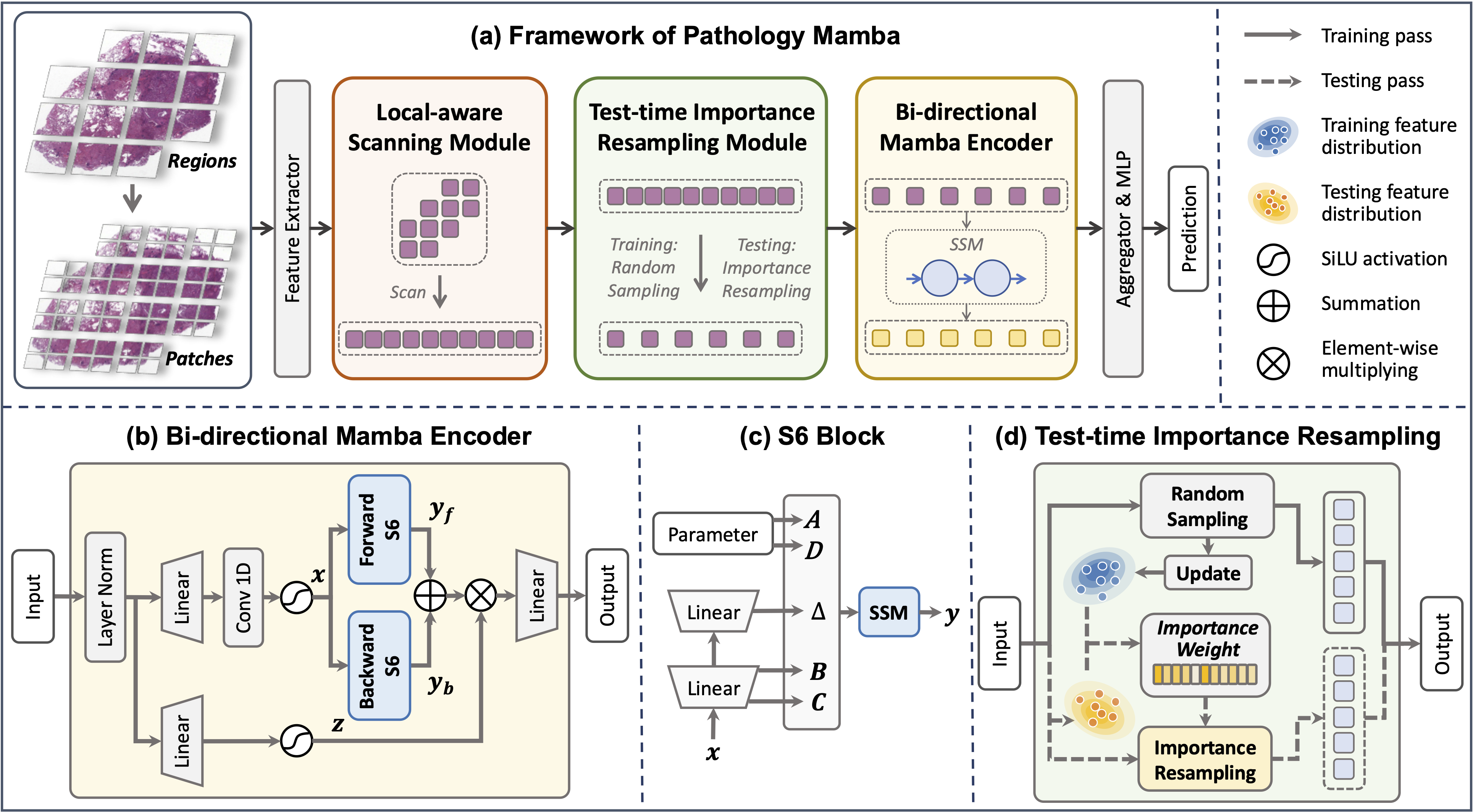
Unleash the Power of State Space Model for Whole Slide Image with Local Aware Scanning and Importance Resampling
Yanyan Huang, Weiqin Zhao, Yu Fu, Lingting Zhu, and Lequan Yu.
IEEE Transactions on Medical Imaging, 2024
This paper introduces a new Pathology Mamba (PAM) for more accurate and robust WSI analysis. It includes three carefully designed components to tackle the challenges of enormous image size, the utilization of local and hierarchical information, and the mismatch between the feature distributions of training and testing during WSI analysis.
-
IEEE TMITAD-Graph: Enhancing Whole Slide Image Analysis via Task-Aware Subgraph Disentanglement, Fuying Wang, Jiayi Xin, Weiqin Zhao, Yuming Jiang, Yeung, Maximus, Liangsheng Wang, Lequan Yu. IEEE Transactions on Medical Imaging. -
MICCAI 2025MoST-IG: Morphology-Guided Spatial Transcriptomics Integration via Visual-Genomic Graph Optimal Transport, Yu Liting, Ma tao, Weiqin Zhao, Zhuo Liang, Lequan Yu. International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), 2025 -
MICCAI 2025HyperPath: Knowledge-Guided Hyperbolic Semantic Hierarchy Modeling for WSI Analysis Peixiang Huang, Yanyan Huang, Weiqin Zhao, Junjun He, Lequan Yu. International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), 2025 -
ISBI 2023Transformer-based Multimodal Fusion for Survival Prediction by Integrating Whole Slide Images, Clinical, and Genomic Data, Yihang Chen, Weiqin Zhao, Lequan Yu. IEEE International Symposium on Biomedical Imaging 2023. -
MLCB 2024ST-200K: A Large-scale Paired Dataset for Histology Imaging and Spatial Transcriptomics, Zhuo Liang$^{*}$, Weiqin Zhao$^{*}$, Fuying Wang, Yuanhua Huang, Lequan Yu. Machine Learning in Computational Biology 2024. -
MLCB 2022Transformer-based Multimodal Fusion for Survival Prediction from Whole Slide Images, Yihang Chen, Weiqin Zhao, Lequan Yu.. Machine Learning in Computational Biology 2022. -
JAILarge-scale self-normalizing neural networks, Zhaodong Chen$^{*}$, Weiqin Zhao$^{*}$, Lei Deng, Yufei Ding, Qinghao Wen, Guoqi Li, Yuan Xie. Journal of Automation and Intelligence. -
ICCRD 2020Aphasia Treatment Assistant System Based on Recommendation and Generation, Lei Li, Jiadong Wang, Rong Ding, Weiqin Zhao, Xuyang Wang, Su Huo. International Conference on Computer Research and Development 2020. -
DSPRApproximate Error Estimation based Incremental Word Representation Learning, Hao Peng, Lin Liu, Liya Ma, Weiqin Zhao, Hongyuan Ma, Long Yuntao. Data Science and Pattern Recognition.
📝 Preprints
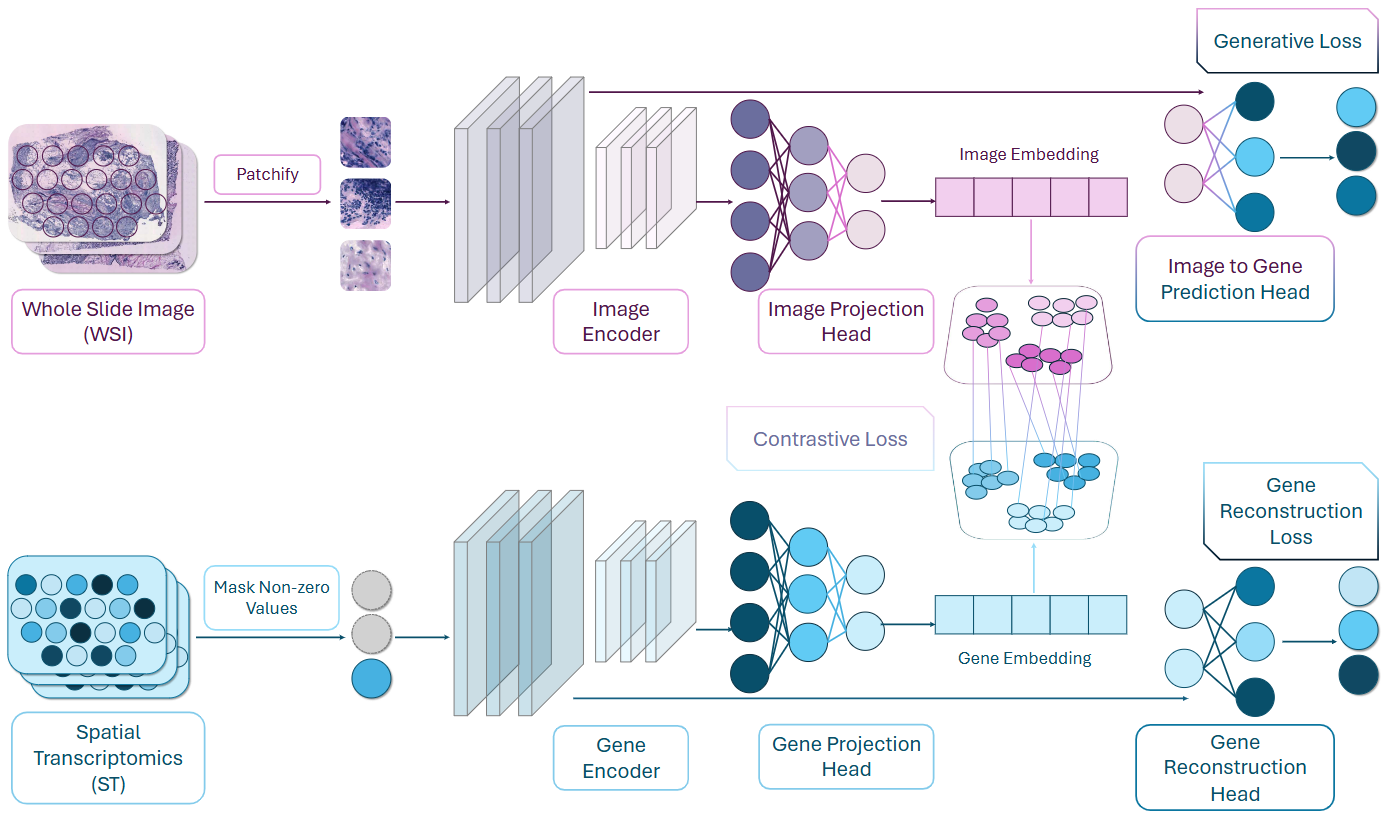
BRIDGE: A Cross-organ Foundation Model for Bridging Histology Imaging and Spatial Transcriptomics
Zhuo Liang$^{*}$, Weiqin Zhao$^{*}$, Fuying Wang, Yuanhua Huang, Lequan Yu.
Present a multi-organ foundation model pre-trained on over 600,000 image-gene paired profiles from 13 organs with various sequencing techniques, to connect cross-organ morphology and genomics for cost-effective histology analysis. Without fine-tuning, BRIDGE enables accurate gene inference for over 80 biomarker genes across 10 human organs, enhances survival analysis for six different types of cancer by using histopathology images from TCGA cohorts and shows potential as a cost-efficient alternative to bulk RNA sequencing.
Journal SubmissionST-Diffusion: A Universal Conditional Diffusion Framework for Spatial Transcriptomics Modeling, Tao Ma, Lingting Zhu, Weiqin Zhao, Jindong Jiang, Lequan Yu.
🎓 Educations
- 2021.09 - 2025.08, Ph.D., School of Computing and Data Science, The University of Hong Kong, Hong Kong SAR, China.
- 2017.09 - 2021.06, B.S., School of Computer Science and Engineering, Beihang Univeristy, Beijing, China.
- 2011.09 - 2017.06, Chengdu Foreign Languages School, Cheng Du, China.
💼 Services
Program Committee
- Program Committee of Machine Learning in Computational Biology (MLCB 2025)
Conference Reviewers
- International Conference on Learning Representations (ICLR 2025-26)
- IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2024-26)
- International Conference on Computer Vision (ICCV 2025)
- The IEEE/CVF Winter Conference on Applications of Computer Vision (WACV 2026)
- International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2025)
- IEEE International Symposium on Biomedical Imaging (ISBI 2025)
- Machine Learning in Computational Biology (MLCB 2023-24)
- Computer Vision for Automated Medical Diagnosis (CVAMD 2023)
- Women in Medical Image Understanding and Analysis (WiMIUA 2022)
Journal Reviewers
- Nature Method (co-reviewer)
- Nature npj Digital Medicine
- IEEE Transactions on Medical Imaging (IEEE TMI)
- Knowledge-Based Systems
- Scientific Reports
- Frontiers in Oncology
- BMC Bioinformatics
- BMC Digital Health
- Journal of Healthcare Informatics Research
🏅 Awards
- AAAI Conference on Artificial Intelligence (AAAI) 2023 Student Scholarship, 2023.
- Department Excellent Research Award, HKU, 2022.
- Annual outstanding student leader of Beihang University, 2019.
- Annual excellent students of Beihang University, 2017-18.
👨🏫 Teaching
- STAT8307 Natural Language Processing and Text Analytics, 2023 and 2025 Spring, HKU.
- STAT3612 Statistical Machine Learning, 2022-24 Fall, HKU.
- STAT8017 Data Mining Techniques, 2022 Spring, HKU.
- STAT2601 Probability and Statistics, 2024 Spring, HKU.











